Citation:
| REPRINT | 2.04 MB |
Abstract:
BACKGROUND:
Although hybridization is thought to be relatively rare in animals, the raw genetic material introduced via introgression may play an important role in fueling adaptation and adaptive radiation. The butterfly genus Heliconius is an excellent system to study hybridization and introgression but most studies have focused on closely related species such as H. cydno and H. melpomene. Here we characterize genome-wide patterns of introgression between H. besckei, the only species with a red and yellow banded 'postman' wing pattern in the tiger-striped silvaniform clade, and co-mimetic H. melpomene nanna.
RESULTS:
We find a pronounced signature of putative introgression from H. melpomene into H. besckei in the genomic region upstream of the gene optix, known to control red wing patterning, suggesting adaptive introgression of wing pattern mimicry between these two distantly related species. At least 39 additional genomic regions show signals of introgression as strong or stronger than this mimicry locus. Gene flow has been on-going, with evidence of gene exchange at multiple time points, and bidirectional, moving from the melpomene to the silvaniform clade and vice versa. The history of gene exchange has also been complex, with contributions from multiple silvaniform species in addition to H. besckei. We also detect a signature of ancient introgression of the entire Z chromosome between the silvaniform and melpomene/cydno clades.
CONCLUSIONS:
Our study provides a genome-wide portrait of introgression between distantly related butterfly species. We further propose a comprehensive and efficient workflow for gene flow identification in genomic data sets.


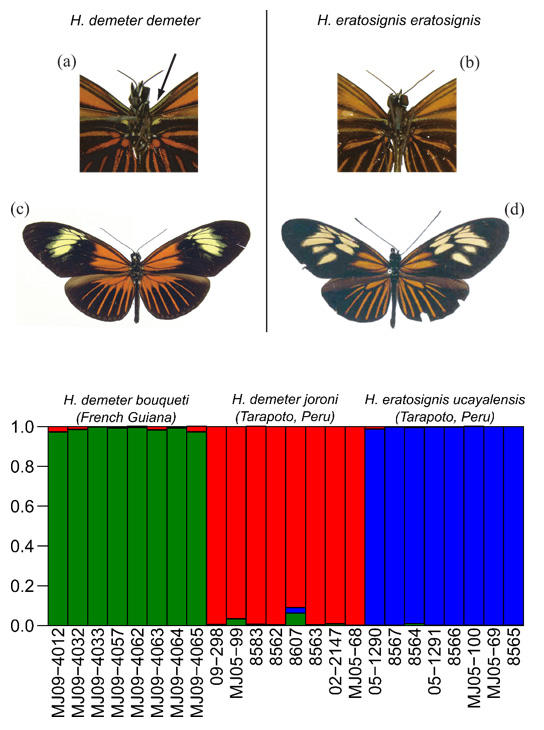
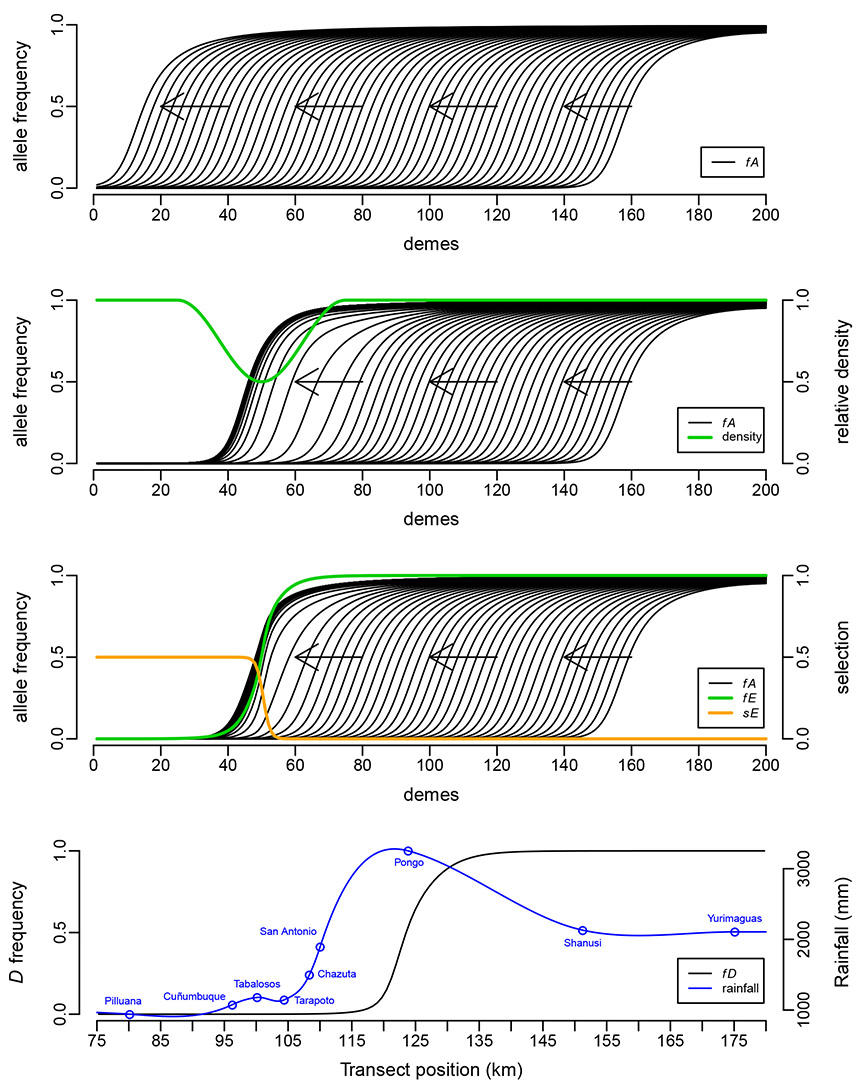
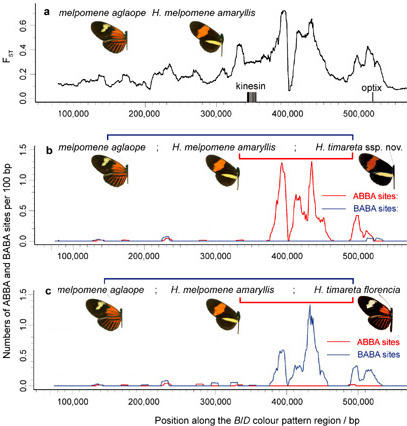
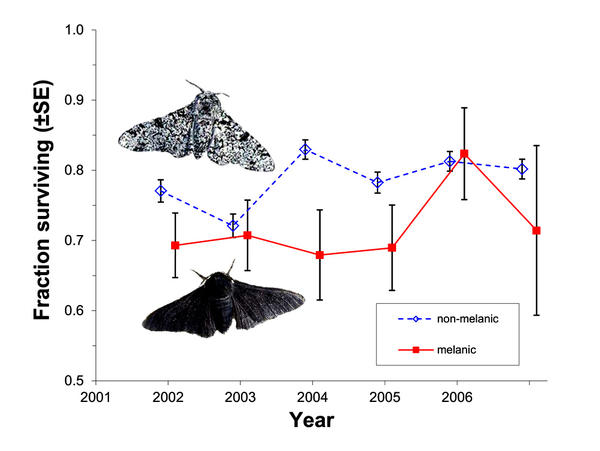 Selection against melanism in UK after the Clean Air Act.
Selection against melanism in UK after the Clean Air Act.